|



Laboratory of David Givol
Research Interests
I. Gene Targeting in Mouse Models
Keywords : Knock in, Gene targeting, FGFR, Achondroplasia
-
Dwarf mice produced by targeting FGFR3
II. Genome-Wide Expression Studies Using Microarrays
Keywords : p53, p73, apoptosis, UVB, skin cancer, stem cells, oligo microarray, DNA chips, clustering, bioinformatics, statistics
- p53 primary and secondary -induced genes
- p73 and p53 specific and common target genes
- Dissection of apoptotic pathways
- UVB specific-induced genes and skin cancer
- Human ES and adult stem cells - Self renewal and pluripotentiality
I. Gene Targeting in Mouse Models
Four fibroblast growth factor receptors (FGFRs) are known, and > 50 mutations in three of them (FGFR1, 2 and 3)
have recently been implicated in congenital skeletal and cranial disorders. Achondroplasia, the most common
form of dwarfism, was shown to be linked to a single point mutation, G380R, in the transmembrane region of FGFR3.
-
Dwarf mice produced by targeting FGFR3
We used gene targeting to introduce the human achondroplasia mutation G380R
into the murine FGFR3 gene. Heterozygotes for this point mutation that carried the neo cassette
were normal whereas neo+ homozygotes had a phenotype similar to FGFR3-deficient mice, exhibiting bone overgrowth.
This was because of interference with mRNA processing in the presence of the neo cassette and represent functional
inactivation of FGFR3. Removal of the neo selection marker by Cre/loxP recombination yielded a dominant dwarf phenotype.
These mice are distinguished by their small size, shortened craniofacial area, hypoplasia of the midface
with protruding incisors, distorted brain case with anteriorly shifted foramen magnum, kyphosis and narrowed
and distorted growth plates in the long bones, vertebrae, and ribs. These experiments demonstrate that
achondroplasia results from a gain-of-FGFR3-function leading to inhibition of chondrocyte proliferation.
Proc. Natl. Acad. Sci. USA, 96, 4455-4460, 1999.
[pdf]
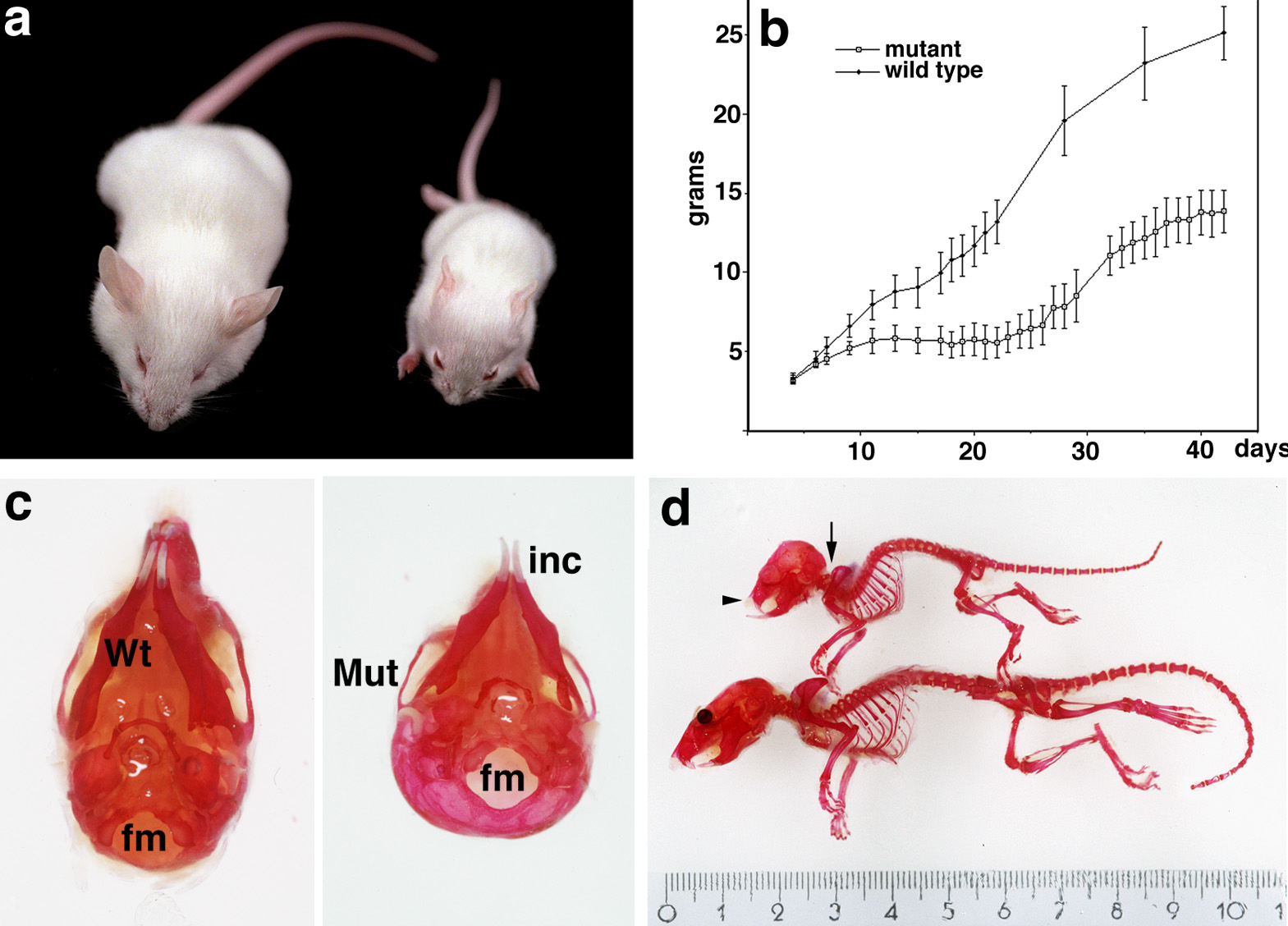
Anatomical defects in FGFR3 knock-in mutants.
a) Dwarfism in 5 weeks old mutant (right) and wild-type littermate control mice (left).
b) Growth curve of the neo- heterozygote and wild-type mice.
c) Comparison of the skull of a wild-type and mutant dwarf mouse showing the change in the foramen magnum (fm) position
and the protruding incisors (inc).
d) Skeletal defects in the dwarf mice. The arrow points to the craniocervical junction and thoracic kyphosis;
the arrowhead shows the facial shortening and overgrowth of incisor teeth.
II. Genome-Wide Expression Studies Using Microarrays
Recently we used the Affymetrix®
oligonucleotide GeneChip® microarray technology to analyze the expression of
thousands of genes in parallel. Total RNA from different samples/experiments was extracted and used to generate target labeled cRNAs
that were hybridized to the latest versions of whole human genome GeneChip® expression microarrays :
HG-U95Av2
(~12,000 mRNA species) and more recently HG-U133A
(~22,000 mRNA species). For all genes in all samples/experiments, the expression signal for each of these genes was determined from
the 'probeset' in use for this gene and by the probe level data analysis software available :
1) signal generated by the Affymetrix®
Microarray Suite Software 5.0
2) or Model Based Expresion Indexes (MBEI) generated by the dChip 1.3 package
recently developed in Wing Wong lab from the Harvard School of Public Health.
Clustering analyses were performed by using the
Supra Paramagetic Clustering (SPC) algorithm,
developped in the lab of Eytan Domany at the Weizmann Institute of Science. More recently, a more sophisticated clustering algorithm
Coupled Two Way Clustering (CTWC), designed to mine gene expression data,
has also been developped and successfully applied. CTWC is an iterative unsupervised procedure, which uses the SPC algorithm as its clustering engine.
The algorithm identifies subsets of genes and samples such that when one of these is used to cluster the other,
stable and significant partitions emerge. The method allows for the detection of small subsets of genes that partition between the samples,
and thereby identifies small number of key genes, which may play a role in a particular biological process.
This procedure avoids the "noise" introduced by other genes, which may mask the signal of the important players.
-
p53-inducible genes
The transcriptional program regulated by the tumor suppressor p53 was analysed using oligonucleotide
microarrays. A human lung cancer cell line (H1299) that expresses the temperature sensitive murine p53 was
utilized to quantitate mRNA levels of various genes at different time points after shifting the temperature to
32C. Inhibition of protein synthesis by cycloheximide (CHX) was used to distinguish between primary and
secondary target genes regulated by p53. Cluster analysis of these data using the super paramagnetic
clustering (SPC) algorithm demonstrate that the primary genes can be distinguished as a single cluster among a
large pool of p53 regulated genes. This procedure identified additional genes that co-cluster with the
primary targets and can also be classified as such genes. The results suggest that p53 activates concerted
opposing signals and exerts its effect through a diverse network of transcriptional changes that collectively
alter the cell phenotype in response to stress.
Oncogene (2001b) 20, 2225 - 2234.
[pdf]
-
p73 and p53 specific and common target genes
Another two p53 and p73 inducible cell lines based on the ecdysone (muristerone) system have been established from the same H1299 cells
(devoid of endogenous p53) as described above that now allow us to detect new p73 and p53 specific and common target genes.
The newly discovered p53 family member, p73, has a striking homology to p53 in both sequence and modular
structure. Ectopic expression of p73 promotes transcription of p53 target genes and recapitulates the most characterized
p53 biological effects such as growth arrest, apoptosis, and differentiation. These phenotypes suggest different biological
activities mediated by p53 and p73 that might reflect activation of specific sets of target genes.
Here, we have analyzed the gene expression profile of H1299 cells after p73alpha or p53 activation.
Our results indicate that p73alpha and p53 activate both common and distinct groups of genes.
Functional classification of these genes reveals that they are involved in many aspects of cell function ranging
from cell cycle and apoptosis to DNA repair.
Furthermore, we report that some of the upregulated genes are directly activated by p73alpha or p53.
J. Biol Chem. (2002) 277, 43359-68.
[pdf]
-
Dissection of apoptotic pathways
Activation of Apaf-1 by p53 :
To study genes activated in the p53 induced apoptotic process, we used a mouse myeloid leukemic cell line (LTR6)
expressing the temperature-sensitive p53 (val135) that undergoes apoptosis upon shifting the temperature to 32C.
Cluster analysis of the p53-regulated genes indicate a pattern of early and late induced sets of genes.
Functional classification of these genes reveals that they are involved in many aspects of cell function, in
addition to growth arrest and apoptosis. Comparison of p53 regulated gene expression profile in LTR6 cells to
that of a human lung cancer cell line (H1299) that undergoes growth arrest but not apoptosis demonstrates
that only 15% of the genes are common to both systems. This observation supports the presence of two distinct
transcriptional programs in response to p53 signaling, one leading to growth arrest and the other to apoptosis.
The proapoptotic genes induced only in LTR6 cells like Apaf-1, Sumo-1 and gelsolin among others may suggest
a possible explanation for apoptosis in LTR6 cells.
Oncogene (2001a) 20, 3449 - 3455.
[pdf]
Inhibition of p53-induced apoptosis by IL-6 :
More recently, we also have analyzed the mechanism of inhibition of wild-type p53-induced apoptosis by the cytokine interleukin-6.
Using DNA microarray and clustering of expressed genes we have analyzed the mechanism of inhibition of wild-type p53-induced apoptosis
by the cytokine interleukin-6 (IL-6) and the calcium mobilizer thapsigargin (TG). Clustering analysis of 1786 genes,
whose expression level changed after activation of wild-type p53 in the absence or presence of IL-6 or TG,
showed that these compounds did not cause a general inhibition of the ability of p53 to upregulate or downregulate gene expression.
Expression of p53-targets implicated as mediators of p53-induced apoptosis were also not affected by IL-6 or TG.
These compounds can thus bypass the effect of wild-type p53 on gene expression to suppress apoptosis.
IL-6 and TG activated different p53-independent pathways of gene expression that include upregulation of antiapoptotic genes and downregulation
of proapoptotic genes. IL-6 and TG also activated different differentiation-associated genes.
The ability of compounds such as cytokines and calcium mobilizers to inhibit p53-mediated apoptosis without generally inhibiting the function of p53>
as a transcriptional activator or repressor, can facilitate tumor development and tumor resistance to radiation and chemotherapy in cells
that retain wild-type p53.>
Proc. Natl. Acad. Sci. USA (2003) in press.
[pdf]
-
UVB specific-induced genes and the induction of
skin cancer
To gain insight into the transformation of epidermal cells into Squamous Carcinoma Cells (SCC),
we compared the response to ultraviolet B radiation (UVB) of Normal Human Epidermal Keratinocytes (NHEK)
to their transformed counterpart, SCC, using biological and molecular profiling.
The major group of upregulated genes in keratinocytes fall into three categories:
(i) antiapoptotic and cell survival factors, including chemokines of the CXC/CC subfamilies, growth factors, and pro-inflammatory mediators,
(ii) DNA repair-related genes, and (iii) ECM proteases. The major downregulated genes are two potential markers for the maintenance
of keratinocyte stem cells. NHEK were found to be more resistant than SCC to UVB-induced apoptosis and this resistance was mainly due to
the protection from cell death by secreted survival factors. Whereas the response of keratinocytes to UVB involved regulation of key
checkpoint genes (p53, MDM2, p21Cip1, DNp63), as well as antiapoptotic and DNA repair-related genes - no or little regulation of
these genes was observed in SCC. To further analyze these changes, we used a novel unsupervised Coupled Two Way Clustering (CTWC) method
that allowed the identification of groups of genes that clearly partitioned keratinocytes from SCC, including a group of genes whose
constitutive expression levels were similar before UVB. This allowed the identification of discriminating genes not otherwise revealed
by simple static comparison in the absence of UVB irradiation. The implication of the changes in gene profile in keratinocytes for
epithelial cancer is discussed.
Oncogene (2003) 22,2993-3006.
[pdf]
-
Human ES and adult stem cells - Self renewal and pluripotentiality
Human embryonic and adult stemness and and their relationship to cancer (under construction)
DESIGN OF AFFYMETRIX GENECHIP® EXPRESSION ANALYSIS SYSTEM
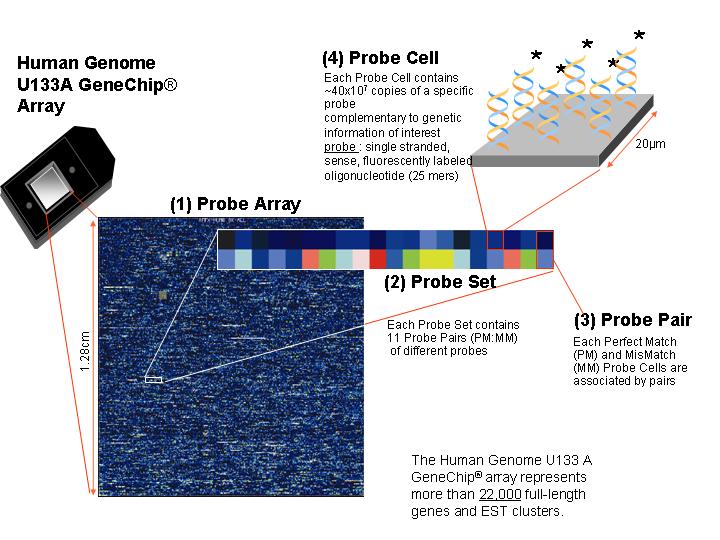
Schematic representation of the Affymetrix GeneChip® expression analysis system in use.
The Probe Array contains ~22,000 Probe Sets on a surface of ~ 1.2 cm2
PROBE SET
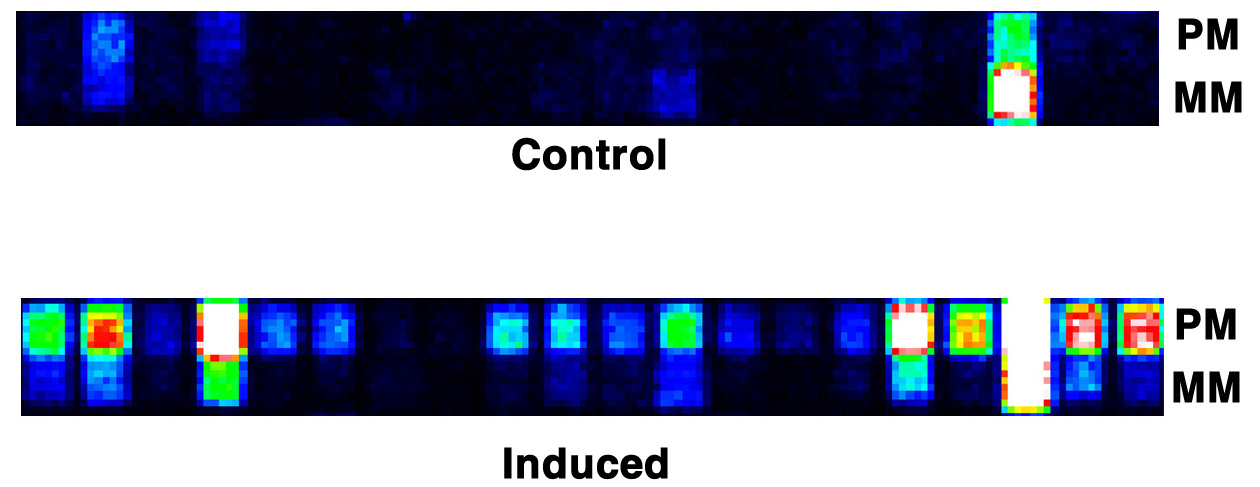
Representation in false color of the differential hybridization (e.g of the p21Waf1 gene)
on two identical Probe Sets (from two different microarray GeneChips®),
between control and induced expression experiments (PM=Perfect Match, MM=MisMatch)
CLUSTERING BY SPC
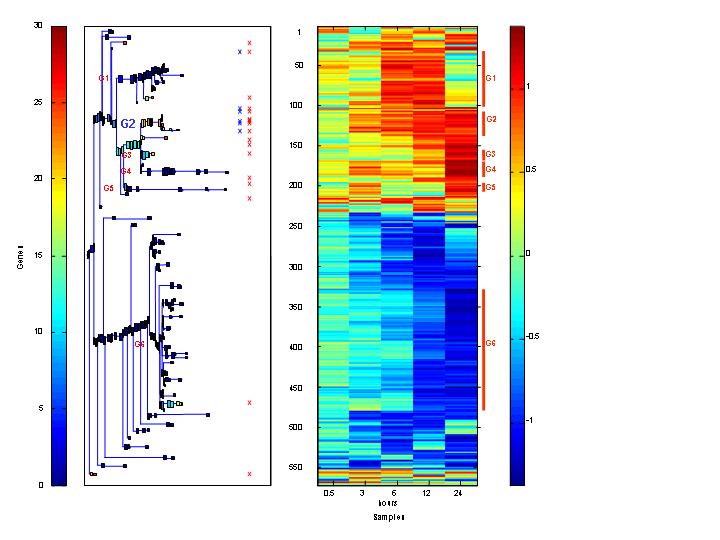
Dendrogram and expression matrix e.g. of gene expression data
CLUSTERING BY CTWC
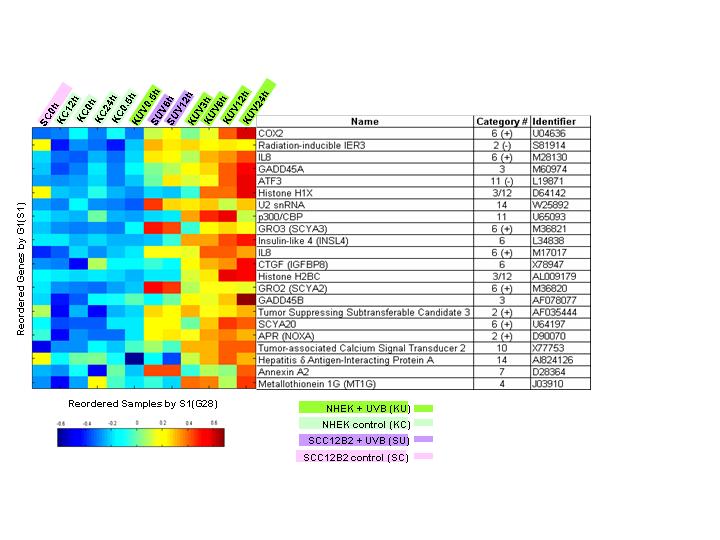
Subset of genes (cluster) determined by CTWC
I. Gene Targeting in Mouse Models
II. Genome-Wide Expression Studies Using Microarrays
TOP
Back to David Givol's Home Page

Weizmann Institute of Science
Department Molecular Cell Biology
76100 Rehovot, Israel
Phone: +972-8-934-3634
Fax: +972-8-934-4125

|






